Software
- SOMbrero (m)
- mixKernel (m)
- SISIR (m)
- nmfprofiler (m)
- treediff (m)
- RNAseqNet (m)
- NiLeDAM (m)
- asterics
- adjclust
- ASICS
- phoenics
- hicream
- Scripts associated to articles
R and Python packages (maintainer)
SOMbrero
SOMbrero is an R package that implements stochastic self-organizing maps (SOM) variants for numeric and non numeric datasets.Project website: http://sombrero.clementine.wf/
Project repositories: Forge INRAE and Github
SOMbrero comes with a Web User Interface (using shiny) that can be used by typing the command line:
sombreroWUI() 
If you are using SOMbrero, please cite:
- Villa-Vialaneix N. (2017) Stochastic self-organizing map variants with the R package SOMbrero. Proceedings of the 12th International Workshop on Self-Organizing Maps and Learning Vector Quantization, Clustering and Data Visualization (WSOM 2017), 1-7.
- Olteanu M., Villa-Vialaneix N. (2015) On-line relational and multiple relational SOM. Neurocomputing, 147, 15-30. DOI: 10.1016/j.neucom.2013.11.047
- Mariette J., Rossi F., Olteanu M., Villa-Vialaneix N. (2016) Accelerating stochastic kernel SOM. XXVth European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning (ESANN 2017), Verleysen M. (ed), i6doc, Bruges, Belgium, 269-274.
mixKernel
mixKernel is an R package that provides methods to combine kernels for unsupervised exploratory analysis. Different solutions are implemented to compute a meta-kernel, in a consensus way or in a way that best preserves the original topology of the data. mixKernel also integrates kernel PCA to visualize similarities between samples in a non linear space and from the multiple source point of view. Functions to assess and display important variables are also provided in the package.Project website: http://mixkernel.clementine.wf/
Project repository: Forge INRAE
If you are using mixKernel, please cite:
- Mariette, J., Villa-Vialaneix, N. (2017) Unsupervised multiple kernel learning for heterogeneous data integration. Bioinformatics, 34(6), 1009-1015. DOI: 10.1093/bioinformatics/btx682
- Brouard, C., Mariette, J., Flamary, R., Vialaneix, N. (2022) Feature selection for kernel methods in systems biology. NAR Genomics and Bioinformatics, 4, lqac014. DOI: 10.1093/nargab/lqac014
SISIR
SISIR is an R package that implements the sparse interval sliced inverse regression. It also provides an implementation of standard ridge and sparse SIR for high dimensional data.Project repository: Forge INRAE
If you are using SISIR, please cite:
- Picheny V., Servien R., Villa-Vialaneix N. (2016) Interpretable sparse SIR for digitized functional data. Statistics and Computing, 29, 255-267. DOI: 10.1007/s11222-018-9806-6
- Servien, R., Vialaneix, N. (2024) A random forest approach for interval selection in functional regression. Statistical Analysis and Data Mining, 17, e11705. DOI: 10.1002/sam.11705
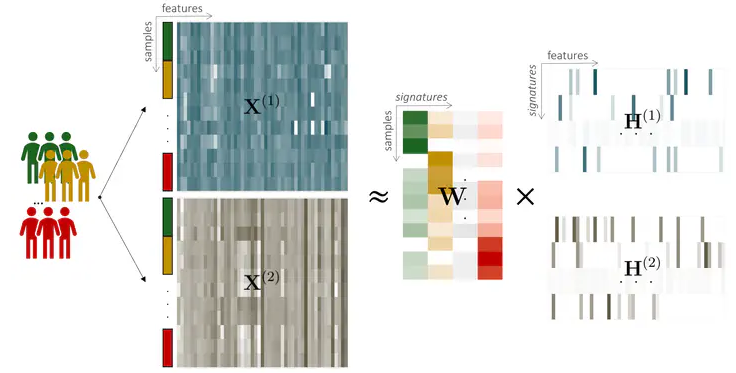
NMFProfiler
NMFProfiler is a Python package (available on PyPi) to perform multi-omics integration for samples stratified in groups. It is based on a supervised version of the NMF.Project repository: Forge INRAE
If you are using NMFProfiler, please cite:
- Mercadié, A., Gravier, É., Josse, G., Fournier, I., Viodé, C., Vialaneix, N., & Brouard, C. (2025) NMFProfiler: A multi-omics integration method for samples stratified in groups. Bioinformatics, 41(2), btaf066. DOI: 10.1093/bioinformatics/btaf066
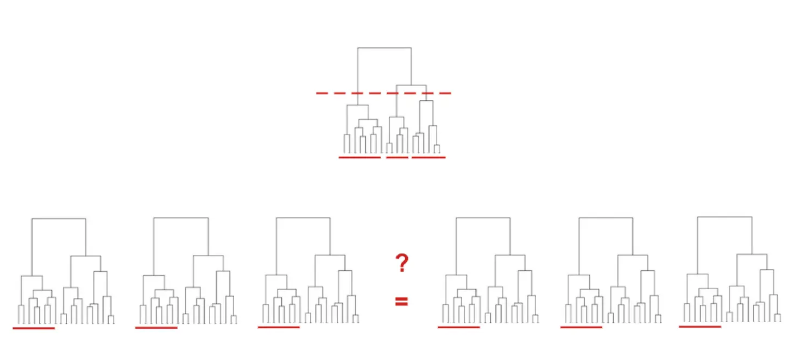
treediff
treediff performs tests to detect differences in structure between families of trees: The method is based on cophenetic distances and aggregated Student’s tests. The package includes a wrapper to deal with Hi-C data (chromatine conformation data based on high-thoughput sequencing).Project repository: Forge INRAE
If you are using treediff, please cite:
- Neuvial, P., Randriamihamison, N., Chavent, M., Foissac, S., Vialaneix, N. (2024) A two-sample tree-based test for hierarchically organized genomic signals. Journal of the Royal Statistical Society, Series C, 73(3), 774-795. DOI: 10.1093/jrsssc/qlae011
RNAseqNet
RNAseqNet is an R package that implements network inference with log-linear Poisson Graphical Model. Hot-deck multiple imputation method can be used to improve the reliability of the inference with an auxiliary dataset.Project repository: Forge INRAE
If you are using RNAseqNet, please cite:
- Imbert, A., Valsesia, A., Le Gall, C., Armenise, C., Gourraud, P.A., Viguerie, N. and Villa-Vialaneix, N. (2017) Multiple hot-deck imputation for network inference from RNA sequencing data. Bioinformatics, 34(10), 1726-1732. DOI: 10.1093/bioinformatics/btx819

NiLeDAM
NiLeDAM is an R package for dating monazites from measurements of triplets (U,Th,Pb) obtained by a microprobe technique. The implemented method is the one described in the article (Montel et al., 1996) and the package has been developed from the original programs made in basic language by Jean-Marc Montel (École Nationale Supérieure de Géologie, France). An example, kindly provided by Anne-Magali Seydoux-Guillaume is included in the package and is the one described in the article (Seydoux-Guillaume et al., 2012).Monazite image by Aangelo (personal work) - licence GFDL or CC-BY-SA-3.0-2.5-2.0-1.0, via Wikimedia Commons
Project repository: Github
If you are using NiLeDAM, please cite:
- Villa-Vialaneix, N., Montel, J.M., Seydoux-Guillaume, A.M. (2013) NiLeDAM: Monazite Datation for the NiLeDAM team. R package version 0.1.
- Montel, J.M., Foret, S., Veschambre, M., Nicollet, C., Provost, A. (1996) Electron microprobe dating of monazite. Chemical Geology, 131, 37-53.
- Seydoux-Guillaume, A.M., Montel, J.M., Bingen, B., Bosse, V., de Parseval, P., Paquette, J.L., Janots, E., Wirth, R. (2012) Low-temperature alteration of monazite: fluid mediated coupled dissolution-precipitation, irradiation damage and disturbance of the U-Pb and Th-Pb chronometers. Chemical Geology, 330-331, 140-158.
- Laurent, A.T., Seydoux-Guillaume, A.M., Duchene, S., Bingen, B., Bosse, V., Datas, L. (2016) Sulphate incorporation in monazite lattice and dating the cycle of sulphur in metamorphic belts. Contributions to Mineralogy and Petrology, 171, 94.
Other software contributions (as author)

asterics
asterics is an web application designed to facilitate statistical and integrative omics data analysis.Maintainer: Élise Maigné
If you are using adjclust, please cite:
- Maigné, É., Noirot, C., Henry, J., Adu Kesewaah, Y., Badin, L., Déjean, S., Guilmineau, C., Krebs, A., Mathevet, F., Segalini, A., Thomassin, L., Colongo, D., Gaspin, C., Liaubet, L., & Vialaneix, N. (2023) ASTERICS: A Simple Tool for the ExploRation and Integration of omiCS data. BMC Bioinformatics, 24, 391. DOI: 10.1186/s12859-023-05504-9
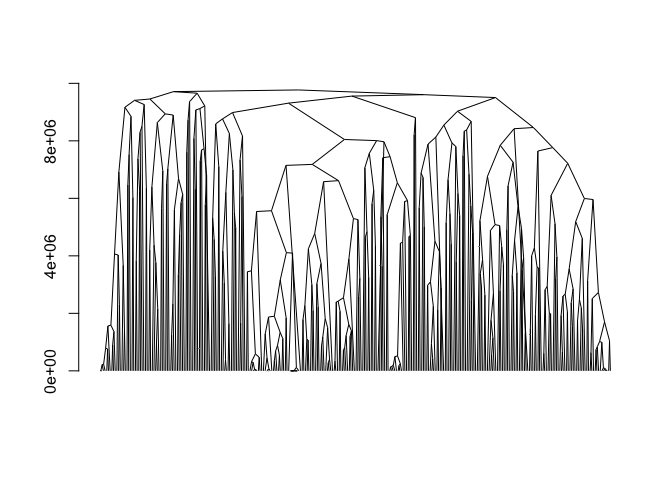
adjclust
adjclust is an R package that implements a constrained version of hierarchical agglomerative clustering, in which each observation is associated to a position, and only adjacent clusters can be merged. Typical application fields in bioinformatics include Genome-Wide Association Studies or Hi-C data analysis, where the similarity between items is a decreasing function of their genomic distance. Taking advantage of this feature, the implemented algorithm is time and memory efficient.Maintainer: Pierre Neuvial
Package website: https://pneuvial.github.io/adjclust/ Project repository: Github
If you are using adjclust, please cite:
- Ambroise, C., Dehman, A., Neuvial, P., Rigaill, G., Vialaneix, N. (2019) Adjacency-constrained hierarchical clustering of a band similarity matrix with application to genomics. Algorithms for Molecular Biology, 14, 363-389. DOI: 10.1186/s13015-019-0157-4
- Randriamihamison, N., Vialaneix, N., Neuvial, P. (2020) Applicability and interpretability of Ward's hierarchical agglomerative clustering with or without contiguity constraints. Journal of Classification, 38, 363-389. DOI: 10.1007/s00357-020-09377-y
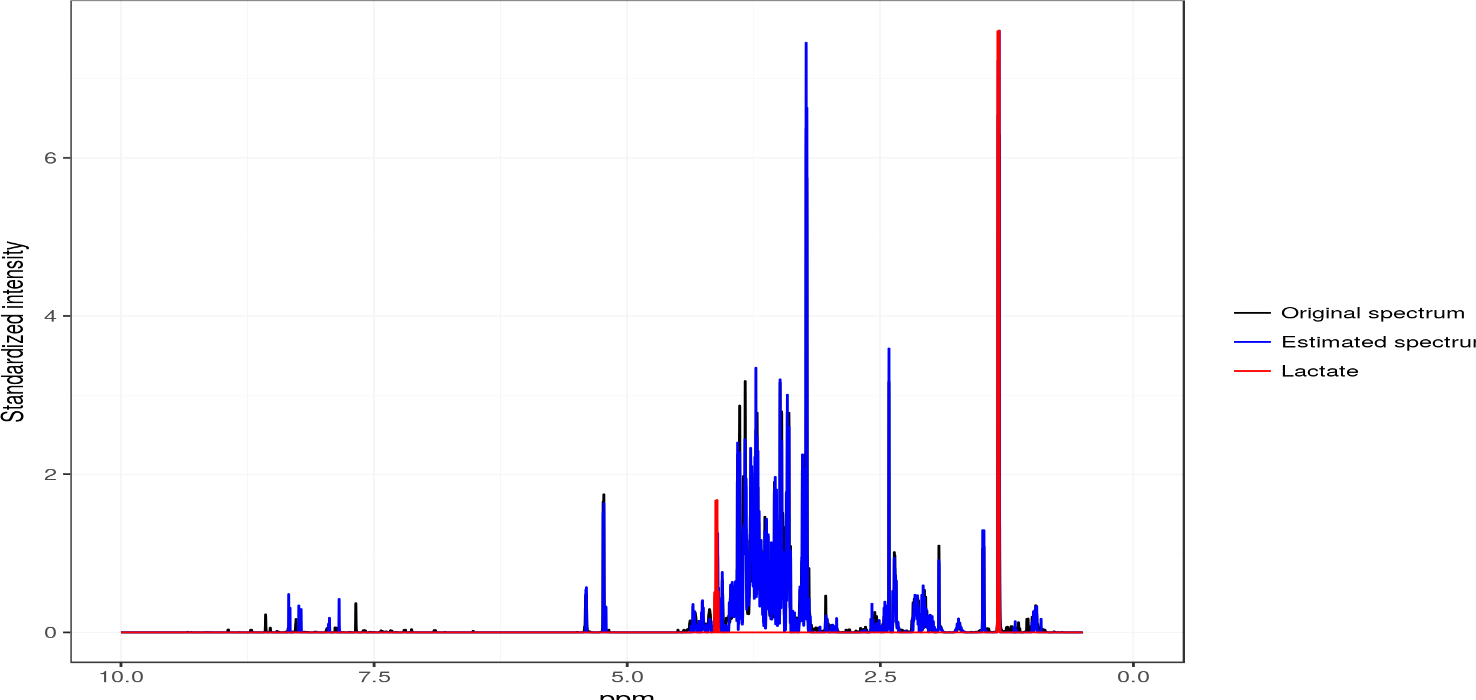
ASICS
ASICS is an R package that quantifies metabolites concentration in a complex spectrum using a set of pure metabolite spectra. The identification of metabolites is performed by fitting a mixture model to the spectra of the library with a sparse penalty.Maintainer: Gaëlle Lefort.
Project repositories: Forge INRAE and
git clone https://git.bioconductor.org/packages/ASICS (Bioconductor source code)If you are using ASICS, please cite:
- Lefort, G., Liaubet, L., Canlet, C., Tardivel, P., Père, M.C., Quesnel, H., Paris, A., Iannuccelli, N., Vialaneix, N., Servien, R. (2019) ASICS: an R package for a whole analysis workflow of 1D 1H NMR spectra. Bioinformatics, 35(21): 4356-4363. DOI: 10.1093/bioinformatics/btz248
- Tardivel, P., Canlet, C., Lefort, G., Tremblay-Franco, M., Debrauwer, L., Concordet, D., Servien, R. (2017). ASICS: an automatic method for identification and quantification of metabolites in complex 1D 1H NMR spectra. Metabolomics, 13(10): 109. DOI: 10.1007/s11306-017-1244-5
phoenics
phoenics is an R package that performs a differential analysis at pathway level based on metabolite quantifications and information on pathway metabolite composition. The method is based on a Principal Component Analysis step and on a linear mixed model. Automatic query of metabolic pathways is also implemented.Maintainer: Camille Guilmineau.
Project repository: Forge INRAE
If you are using ASICS, please cite:
- Guilmineau, C., Tremblay-Franco, M., Vialaneix, N., & Servien, R. (2025) phoenics: a novel statistical approach for longitudinal metabolomic pathway analysis. BMC Bioinformatics, 26, 105. DOI: 10.1186/s12859-025-06118-z

hicream
hicream performs Hi-C data differential analysis based on pixel-level differential analysis and a post hoc inference strategy to quantify signal in clusters of pixels. Clusters of pixels are obtained through a connectivity-constrained two-dimensional hierarchical clustering.Maintainer: Élise Jorge.
Project repository: Forge INRAE
Public repositories associated to articles (scripts, data)

Code (and some data) repositories
Disclaimer! Unlike previous software implementations, these repositories are usually not actively maintained.-
Jorge, É., Foissac, S., Neuvial, P., Zytnicki, M., & Vialaneix, N. (2025) A comprehensive review and benchmark of differential analysis tools for Hi-C data. Briefings in Bioinformatics, 26(2), bbaf074. DOI: 10.1093/bib/bbaf074.
Code (forge INRAE) and Data (recherche.data.gouv) -
Neuvial, P., Randriamihamison, N., Chavent, M., Foissac, S., Vialaneix, N. (2024) A two-sample tree-based test for hierarchically organized genomic signals. Journal of the Royal Statistical Society, Series C, 73(3), 774-795. DOI: 10.1093/jrsssc/qlae011.
Code (forge INRAE) and Data (recherche.data.gouv)
Associated with the R package treediff. -
Servien, R., Vialaneix, N. (2024) A random forest approach for interval selection in functional regression. Statistical Analysis and Data Mining, 17, e11705. DOI: 10.1002/sam.11705.
Code (forge INRAE) and Data (recherche.data.gouv)
Associated with the R package SISIR. -
Brouard, C., Mourad, R., Vialaneix, N. (2024) Should we really use graph neural networks for transcriptomic prediction? Briefings in Bioinformatics, 25(2), bbae027. DOI: 10.1093/bib/bbae027.
Code (forge INRAE) and Data (recherche.data.gouv) -
Marti-Marimon, M., Vialaneix, N., Lahbib-Mansais, Y., Zytnicki, M., Camut, S., Robelin, D., Yerle-Bouissou, M., Foissac, S. (2021) Major reorganization of chromosome conformation during muscle development in pig. Frontiers in Genetics, 12, 748239. DOI: https://doi.org/10.15454/DOMEHB.
Code (forge INRAE) and Data (recherche.data.gouv) -
Picheny V., Servien R., Villa-Vialaneix N. (2016) Interpretable sparse SIR for digitized functional data. Statistics and Computing, 29, 255-267. DOI: 10.1007/s11222-018-9806-6.
Code (Github)
Associated with the R package SISIR. -
Marti-Marimon, M., Vialaneix, N., Voillet, V., Yerle-Bouissou, M., Lahbib-Mansais, Y., Liaubet, L. (2018) A new approach of gene co-expression network inference reveals significant biological processes involved in porcine muscle development in late gestation. Scientific Report, 8, 10150. DOI: 10.1038/s41598-018-28173-8.
Code (Github), including some results. -
Genuer, R., Poggi, J.-M., Tuleau-Malot, C., Villa-Vialaneix, N. (2017) Random forests for big data. Big Data Research, 9, 28-46. DOI: 10.1016/j.bdr.2017.07.003.
Code (Github)
